Assisted Machine learning analysis
The goal of this notebook is to create models to understand the influence of static variables on a dynamic variable. In this example we are going to predict the UER, but any other dynamic variable can be predicted.
If you want more explanations, you can find in the documentation a part dedicated to explain the different concepts.
Import
[1]:
import frflib
import sklearn
from frflib.utils.ml_analysis.optimizer import MLDataset
from frflib.utils.ml_analysis.processing import (
make_static_list,
make_pipeline,
make_dict_preprocessing,
ONE_HOT_ENCODER,
TARGET_ENCODER,
REGROUP_ENCODER,
XYSCALER,
MINMAX,
STANDARD_SCALER,
UNCHANGED,
)
from frflib.utils.ml_analysis.analysis import (
find_best_model_param,
model_analysis,
fit_model,
build_model_summary,
)
from frflib.utils.ml_analysis.simulation import (
get_truncated_normal,
get_percentile,
discretize,
check_enough_data,
generate_df_simulation,
)
from sklearn.model_selection import train_test_split
from frflib.data_class.input_data import InputData
from frflib.forecast_methods import forecast_wf
from frflib.plots.plotly_ import plot_scatter
import plotly.express as px
ForecastWF = forecast_wf.ForecastWF
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import plotly.figure_factory as ff
import plotly.express as px
import os
import inspect
from sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error
/env/lib/python3.8/site-packages/category_encoders/target_encoder.py:92: FutureWarning: Default parameter min_samples_leaf will change in version 2.6.See https://github.com/scikit-learn-contrib/category_encoders/issues/327
warnings.warn("Default parameter min_samples_leaf will change in version 2.6."
/env/lib/python3.8/site-packages/category_encoders/target_encoder.py:97: FutureWarning: Default parameter smoothing will change in version 2.6.See https://github.com/scikit-learn-contrib/category_encoders/issues/327
warnings.warn("Default parameter smoothing will change in version 2.6."
Load data
[2]:
PATH_FRFLIB = os.path.dirname(inspect.getsourcefile(frflib))
PATH_TO_DATA = os.path.join(PATH_FRFLIB, "sample_data")
project_id = "01"
wf_name = "dataset_01_simple_wf"
WF_PATH = os.path.join(PATH_TO_DATA, f"{wf_name}.hdf")
DATA_PATH = os.path.join(PATH_TO_DATA, f"dataset_{project_id}.hdf")
[3]:
field_data = InputData.load_hdf(DATA_PATH)
df_st = field_data.df_static
wf = ForecastWF.load_from_hdf(WF_PATH, field_data)
field_data._compute_indicators()
wf.enrich_data()
df_static = field_data.df_static
df_static
[3]:
| production_name | wellbore | reservoir | sub_group | ptr_main_layer_total | ptr_main _layer | ptr_secondary_layer | cluster | avg_poro_vv | avg_vcl_vv | ... | z_bh_tvdss_m.1 | top_res_sstvd_m | top perfo tvd_ss_ft | z_wh_tvdss_m.1 | sensor_depth_tvdss | init_res_pr_bar | init_depletion_pr_bar | start_up_date | year_start | first_prod_days | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wellname | |||||||||||||||||||||
| well_0 | WA12 | WA12 | Deltaic | B | B22 | B2 | None | WA | 0.54 | 0.39 | ... | 1077.26 | 1036.50 | 1063.72 | 497.48 | 962.016 | 519.86 | 4.04 | 2006-12-01 | 2006 | 2191 |
| well_1 | JD28 | JD28 | Fluvial | D | D2 | D2 | None | JD | 0.77 | 0.23 | ... | 1115.73 | 1003.26 | 1087.60 | 332.30 | 982.500 | 550.00 | 1.01 | 2014-05-01 | 2014 | 4899 |
| well_10 | SA29 | SA29_st1 | Fluvial | D | D3 | D1 | D2 | SA | 0.64 | 0.39 | ... | 1295.13 | 1231.86 | 1308.72 | 626.28 | 1209.727 | 592.00 | 4.63 | 2011-08-01 | 2011 | 3895 |
| well_100 | RA08 | RA08 | Fluvial | D | D3 | D3 | None | RA | 0.82 | 0.26 | ... | 1511.14 | 1507.18 | 1444.31 | 556.99 | 1329.310 | 680.00 | 6.41 | 2009-03-01 | 2009 | 3012 |
| well_101 | JB32 | JB32_st1 | Fluvial | D | D2 | D1 | None | JB | 0.70 | 0.32 | ... | 1171.23 | 1105.51 | NaN | 375.00 | NaN | 490.00 | 8.07 | 2017-01-01 | 2017 | 5875 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| well_93 | YC13 | YC13 | Deltaic | B | B22 | B2 | None | YC | 0.68 | 0.30 | ... | 1166.93 | 1093.93 | 1131.87 | 505.13 | 1021.972 | 540.00 | 4.29 | 2010-09-01 | 2010 | 3561 |
| well_94 | VD17 | VD17 | Deltaic | B | B22 | B2 | None | VD | 0.64 | 0.40 | ... | 1121.12 | 1049.24 | 1101.96 | 511.04 | 1039.875 | 570.00 | 0.94 | 2003-03-01 | 2003 | 820 |
| well_95 | YA18 | YA18_st2 | Fluvial | E | E1 | E1 | None | YA | 0.51 | 0.41 | ... | 1452.83 | 1463.29 | 1480.16 | 479.44 | 1352.500 | 670.00 | 5.85 | 2007-08-01 | 2007 | 2434 |
| well_97 | P136 | P136 | Fluvial | D | D3 | D3 | None | P1 | 0.74 | 0.30 | ... | 1301.07 | 1309.64 | 1329.15 | 561.55 | 1249.500 | 657.00 | 2.37 | 2013-08-01 | 2013 | 4626 |
| well_99 | P302 | P302_st1 | Fluvial | D | D3 | D3 | None | P3 | 0.73 | 0.22 | ... | 1320.42 | 1311.99 | 1334.13 | 535.87 | 1246.300 | 610.00 | 5.68 | 2016-08-01 | 2016 | 5722 |
176 rows × 38 columns
I. Create Model
The first part is to create a model. We are going to choose a model, create a pipeline with some transformations and then train it.
Parameters
The goal is to predict a dynamic variable. You can select here the variable to predict and the models to use. You can add multiple models, but it will take more time. The available models are - gradientBoosting - RandomForest - Lasso - Ridge DATASETS = [“train”, “validation”]
def create_df_result(best_model_dict): model_df = pd.DataFrame() for data_set in DATASETS: data = { key: values for key, values in best_model_dict[data_set].items() if type(values) == list } temp = pd.DataFrame(data=data).set_index(“wellname”) temp[“dataset”] = data_set model_df = pd.concat([model_df, temp], axis=0) return model_df
[4]:
target = "oil_cum_mmbbl"
models = ["GradientBoosting", "Lasso", "RandomForest"]
metric = "neg_mean_squared_error"
availables_metrics = sorted(sklearn.metrics.SCORERS.keys())
assert (
metric in availables_metrics
), f"metrics not in sklearn metrics please use one of {availables_metrics}"
[5]:
df_predict = field_data.df_dynamic.groupby("wellname").max()
df = pd.concat([df_predict[target], df_static], axis=1)
Cleaning
In this part we drop the wells that didn’t start to produce.
[6]:
df = df.loc[df[target].notnull(), :]
Preprocessing
We have implemented some transformers for basic preprocessing. By default we use the targetEncoder for categorical variable and minmax for numerical data.
All transformers available Categorical - targetEncoder : Replace the categories of categorical columns by the mean of the target - OneHotEncoder
Numerical - XYscaler : scaler to be used with geographical column (x,y) - StandardScaler - minmax : rescale the data between 0 and 1 - unchanged : apply no rescaling on the data
Below you have access to the default a list with each column the default transformer we apply.
If you want to modify the preprocessing, you can follow the below example. You can do 2 things: - don’t keep the column (see example on wellbore below) - Choose another transformer for a column (see example on reservoir)
[7]:
static_col = make_static_list(df.drop([target], axis=1))
col_drop = [
"gross_lentgh_m",
"x_wh_m",
"y_wh_m",
"z_wh_tvdss_m",
"z_bh_tvdss_m",
"z_bh_tvd_m",
"azim_deg",
"virgin_res_pr_bar",
"last_res_pr_bar",
"z_bh_tvdss_m.1",
"top perfo tvd_ss_ft",
"z_wh_tvdss_m.1",
"sensor_depth_tvdss",
"init_res_pr_bar",
"init_depletion_pr_bar",
"first_prod_days",
"reservoir.1",
"ptr_main_layer_total",
"multi_lateral_flag",
"ptr_main _layer",
"ptr_secondary_layer",
"well_Type",
"production_name",
"top_res_sstvd_m",
"year_start",
"ptr_main _layer",
"net_length_m",
"drill_length_md_m",
#'reservoir',
#'wellbore',
# "avg_vcl_vv",
# "avg_ntg_vv",
# "sub_group",
# "start_up_date",
# "cluster",
#'incl_max_deg',
#'top_res_sstvd_m'
# "x_bh_m",
# "y_bh_m"
]
for col in static_col:
if col["Name"] == "x_bh_m" or col["Name"] == "y_bh_m":
col["Transform"] = XYSCALER
if col["Name"] == "reservoir":
col["Transform"] = ONE_HOT_ENCODER
if col["Name"] == "top_res_sstvd_m":
col["Transform"] = MINMAX
# To choose to delete column from training
if col["Name"] in col_drop:
col["Keep"] = False
print("Use in model")
set(df.columns) - set(col_drop)
Use in model
[7]:
{'avg_ntg_vv',
'avg_poro_vv',
'avg_vcl_vv',
'cluster',
'incl_max_deg',
'oil_cum_mmbbl',
'reservoir',
'start_up_date',
'sub_group',
'wellbore',
'x_bh_m',
'y_bh_m'}
Pipeline
We then create a sklearndf pipeline with the preprocessing options and the selected models.
[8]:
dict_preprocessing = make_dict_preprocessing(static_col)
preprocessor = make_pipeline(dict_preprocessing)
dict_preprocessing
[8]:
{'numerical': {'minmax': ['avg_poro_vv',
'avg_vcl_vv',
'avg_ntg_vv',
'incl_max_deg'],
'normalize_standard': [],
'XYscaler': ['x_bh_m', 'y_bh_m'],
'unchanged': []},
'categorical': {'OneHotEncoder': ['reservoir'],
'targetEncoder': ['wellbore', 'sub_group', 'cluster']},
'date_time': {'date_time_encoder': ['start_up_date']}}
Fits models
Once the preprocessing is done you can launch the training of the models.
[9]:
# fit on models
# metrics available
# sorted(sklearn.metrics.SCORERS.keys())
# columns = [x["Name"] for x in static_col if x["Keep"] == True]
df_train, df_test = train_test_split(df, test_size=0.3, random_state=35)
data = MLDataset(
df_train.drop([target], axis=1),
df_test.drop([target], axis=1),
df_train[target],
df_test[target],
)
[10]:
best_model_name, grid, pipe_processor, optimisation_per_model = find_best_model_param(
data,
preprocessor,
metric=metric,
budget=5,
models=models,
feature_selection=False,
feature_penalisation=False,
)
data_out, model = fit_model(best_model_name, grid, data, pipe_processor)
model_summary = build_model_summary(data_out)
model_summary.update(
{"optimization_metric": metric, "model": best_model_name, "params": grid}
)
(dict_dependence_plot, dict_summuray_plot, shap_values, df_shap) = model_analysis(
data_out, model, best_model_name
)
Default parameter min_samples_leaf will change in version 2.6.See https://github.com/scikit-learn-contrib/category_encoders/issues/327
Default parameter smoothing will change in version 2.6.See https://github.com/scikit-learn-contrib/category_encoders/issues/327
Unlike other reduction functions (e.g. `skew`, `kurtosis`), the default behavior of `mode` typically preserves the axis it acts along. In SciPy 1.11.0, this behavior will change: the default value of `keepdims` will become False, the `axis` over which the statistic is taken will be eliminated, and the value None will no longer be accepted. Set `keepdims` to True or False to avoid this warning.
Default parameter min_samples_leaf will change in version 2.6.See https://github.com/scikit-learn-contrib/category_encoders/issues/327
Default parameter smoothing will change in version 2.6.See https://github.com/scikit-learn-contrib/category_encoders/issues/327
Unlike other reduction functions (e.g. `skew`, `kurtosis`), the default behavior of `mode` typically preserves the axis it acts along. In SciPy 1.11.0, this behavior will change: the default value of `keepdims` will become False, the `axis` over which the statistic is taken will be eliminated, and the value None will no longer be accepted. Set `keepdims` to True or False to avoid this warning.
[11]:
plt.figure(figsize=(12, 8))
for model_name in models:
score_list = optimisation_per_model[model_name]
score = [x[0] for x in score_list]
penalisation = [x[1] for x in score_list]
plt.plot(score, label=f"score {model_name}")
if np.sum(penalisation) > 0:
plt.plot(penalisation, label=f"penalisation {model_name}")
plt.ylim(bottom=0)
plt.legend()
plt.title(f"Optimizer min score = {min(score):.1f} metric : {metric}")
plt.show()
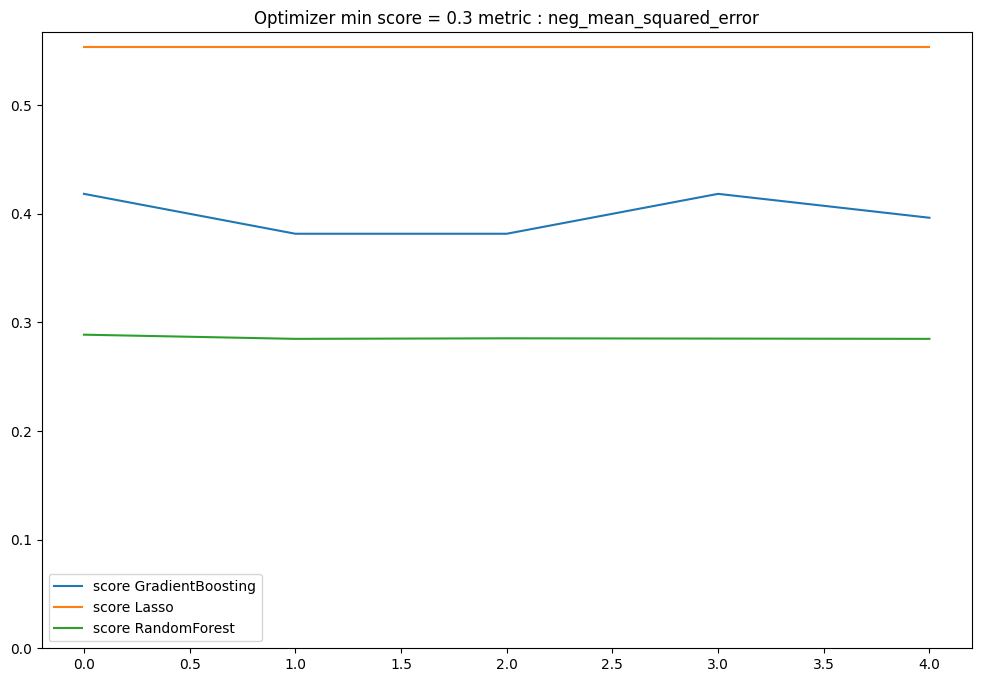
II. Model Analysis
Result
Once the preprocessing is done you can launch the training of the models and then visualise the result of the best model. A good indicator is the r2 square. - r2 > 0.6 : really good r2, you have a strong model - 0.6 > r2 > 0.4 : Acceptable r2 - r2 < 0.4 : Low r2, the model is not accurate enough print(f”Best model {model_summary[‘model’]} :nbsphinx-math:`nParams `= {model_summary[‘params’]}”) df_result = create_df_result(model_summary)
metric_func = {“r2”: r2_score, “mae”: mean_absolute_error, “rmse”: mean_squared_error} result_score = {“train”: {}, “validation”: {}}
for dataset in result_score.keys(): original, pred = ( df_result[df_result.dataset == dataset].target, df_result[df_result.dataset == dataset].predict, ) print(f”{dataset}”) for key, func in metric_func.items(): if key == “rmse”: score = func(original, pred, squared=False) else: score = func(original, pred) result_score[dataset].update({key: score}) print(f”\t{key} = {score:.2f}”)
fig_model = plot_scatter( df_result, “target”, “predict”, color=”dataset”, hover_name=”wellname”, same_scale=True, x_range=[0, 70], y_range=[0, 70], title=”Model with all wells”, idendity=True, ) fig_model.show()
[12]:
# inspection
(
dict_dependence_plots,
dict_summuray_plot,
shap_values,
df_shap,
) = model_analysis(data_out, model, model_name)
Model analysis
Now that we have a model, we are going to make some analysis to understand the influcence of the static variables on the target.
Summuray Plot
This plot gives a summary of the influence and the correlation between the variables and the target. The x-axis is the mean absolute impact on all the wells in term of the target. The variables are sorted according to their impact, and the colour is the correlation (positive or negative with the target).
[13]:
def make_summary_fig(dict_summuray_plot, number_features_plot):
summuray = pd.DataFrame(data=dict_summuray_plot["data"])
summuray = summuray[:number_features_plot][::-1]
fig = px.bar(
summuray,
x="mean_abs",
y="features",
color="correlation",
title=f"Impact on {target}",
orientation="h",
color_continuous_scale="Bluered",
range_color=(-1, 1),
)
return fig
number_features_plot = 30
fig = make_summary_fig(dict_summuray_plot, number_features_plot=15)
fig.show()
[14]:
# This plot shows the impact of a variable on the target. Compare to the first plot, we can explore each variable at a time. On the x-axis you have the values of the selected variable (which is normalised between [0,1]). On the y-axis you have the impact of the feature. This plot allows us to understand the relationship between the target and variable (linear, monotonic etc…).
df_impact = pd.DataFrame(dict_dependence_plots["data"]).set_index("wellname")
# df_impact = pd.concat([df_impact, df], axis=1)
variable = [x[7:] for x in df_impact.columns if x[:6] == "impact"]
for var in variable[0:2]:
if f"{var}" in df_impact.columns:
f = px.scatter(df_impact, var, f"impact_{var}")
f.show()